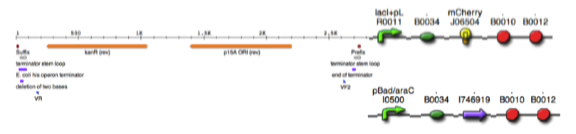
This is an example of how to exchange one gene for another whilst retaining the desired promoter, ribosomal binding site (rbs) and terminator DNA sequences of the gene to be replaced based on Biobrick parts from the Registry of Standard Biological Parts. In this example we will directly replace the gene for Red Fluorescent Protein (RFP), mCherry with the gene for a super folder Green Fluorescent Protein (GFP), whilst keeping the plasmid backbone and regulatory sequences of the gene to be replaced. The process of designing the necessary oligo primers for later Gibson DNA assembly will be illustrated.
Go to the Registry of Standard Biological Parts web address http:partsregistry.org/
and use the search box to recall the relevant Biobrick part.
In this example we are searching for the RFP Biobrick with the part number BBa_J69512.
Having pulled the part page describing the RFP Biobrick we need to download the DNA sequence file in the Genbank format (.gb) using the Tools tab.
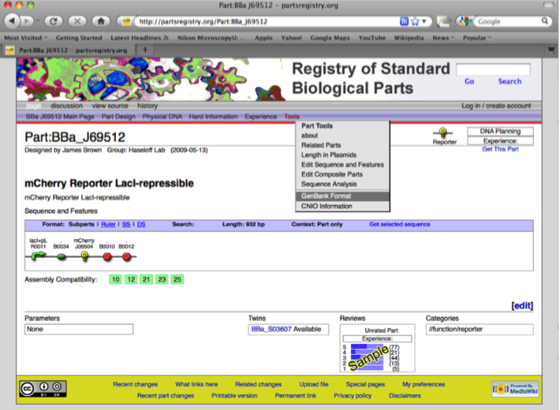
The page downloaded looks like this.
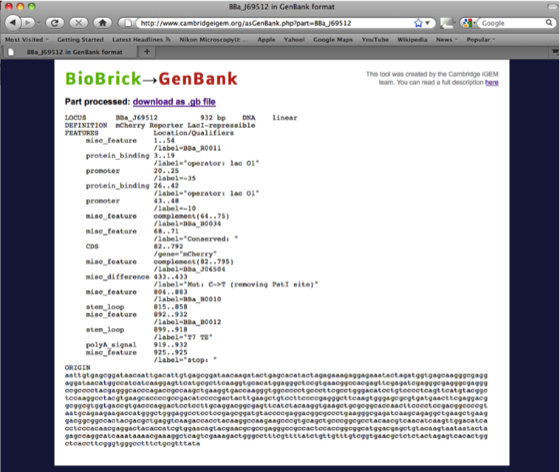
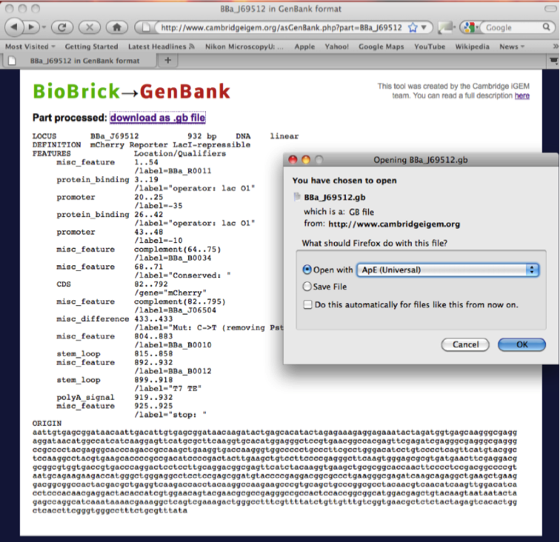
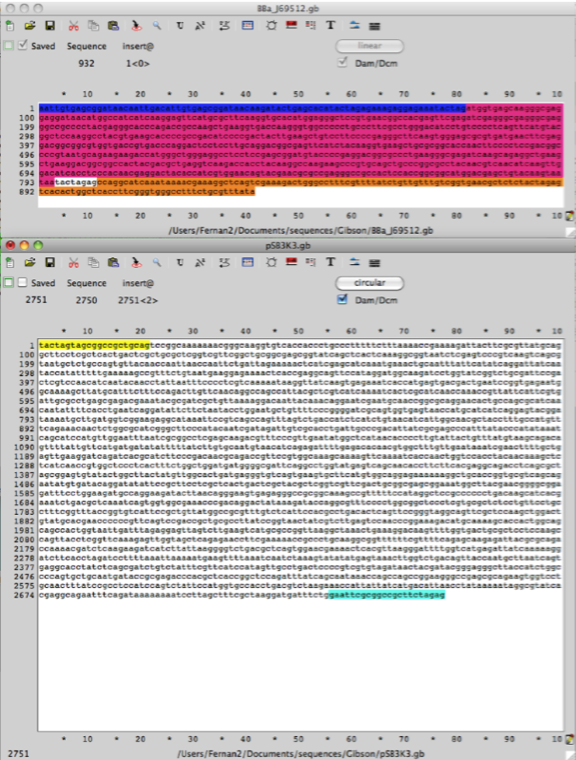
The advantage of the conversion from the Biobrick to Genbank file format is that the annotation of the DNA is retained in Ape. All recognized features (promoters, ribosomal binding sites and sequences of interest are highlighted in colours, which can be edited). These features will help in the design of primers for Gibson assembly.
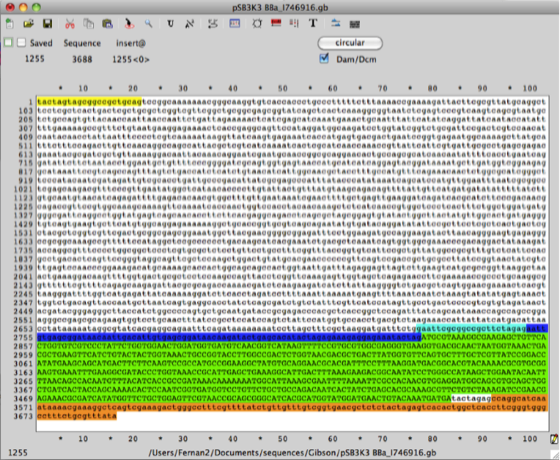
Step 2. Using the same procedure outlined in step 1 create an Ape file for the plasmid pSB3K3 which has the part number (http://partsregistry.org/Part:pSB3K3). Cut & paste the DNA sequence from the RFP (mCherry) BBa_J69512 into the backbone plasmid vector pSB3K3 and use this new sequence as a template to design primers for PCR amplification of the vector sequence including promoter, RBS and terminators flanking BBa_J06504 RFP (mCherry), this part is the gene open reading frame (ORF).
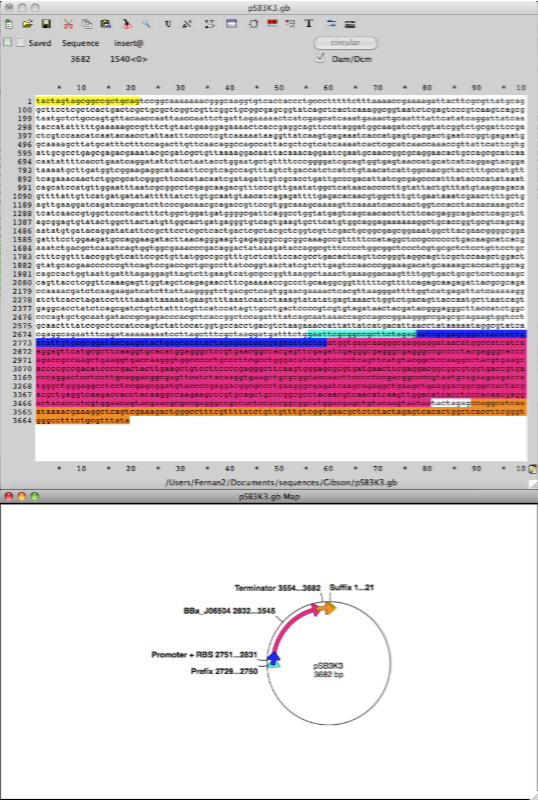
Step 3. Using the same procedure outlined in steps 1 and 2 create an Ape file for the super folder GFP Biobrick BBa_I746916 (http://partsregistry.org/Part:BBa_I746916) then using the Ape file pSB3K3 RFP (mCherry) BBa_J69512 replace the mCherry ORF with the super folder GFP. This will be the in silico template file for designing the oligo primers for Gibson assembly