You will need:
Heating block set to 50ºC
1.5ml eppendorf tubes
Axygen Tips, gel cutting, 1.1mm x 4mm (VWR cat no. 732-1409 for pack/250)
QIAquick gel extraction kit (Qiagen cat no. 28704 for 50 rxns)
Place the agarose gel on a OHP projector sheet, itself on top of the Safe Imager™ 2.0 Blue-Light Transilluminator. (The OHP sheet is there to protect the surface of the transilluminator).
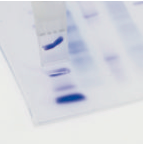
A very quick and easy way to cut DNA bands from agarose gels is to use gel cutting tips available from Axygen. These tips come in two sizes and fit most 1ml pipettors. Place the tip on the end of the pipettor and displace the pipette plunger fully and hold that position.
Turn on the transilluminator and place the tip over the DNA band to be extracted. Press the tip down into the gel and slowly raise the pipette plunger. You may need to repeat this process to capture the entire DNA band.
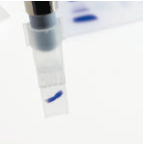
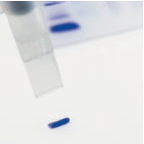
The gel slice is sucked up into the cutting tip and should be transferred to a 1.5ml eppendorf tube. (It may be necessary to sharply tap on the bench the gel cutting tip with the eppendorf tube to remove all of the gel slice).
There are a number of extraction protocols for the removal and purification of DNA from agarose gel. The QIAquick® protocol from Qiagen QIAquick® Spin Handbook (version 2008) is described below.
Measuring DNA using a NanoDrop Spectrophotmeter (http://www.nanodrop.com) - or equivalent, such as the GE NanoVue

Nanodrops are spectrophotometers that have the ability to measure DNA in as little as 0.5-2µl of sample up to 15,000ng/µl. The sample to be measured is place on a pedestal which has a fibre optic cable (the receiving fiber) embedded inside. A second fiber optic cable (the source fiber) is then brought into contact with the sample causing the liquid to bridge the gap between the ends of the two fibers. A pulsed xenon flash lamp is used as a light source and a spectrophotometer using a CCD array analysis records the light that has passed through the sample.
DNA is quantified using a modification of the Beer-Lambert equation allowing the use of a factor of ng-cm/microliter.
c= (A * E)/b
c= the nucleic acid concentration in ng/microliter
A= the absorbance in AU
E= the wavelength-dependent extinction coefficient in ng-cm/microliter
b= the path length in cm
The extinction coefficients for nucleic acids are:
Double stranded DNA: 50ng-cm/µl
Single stranded DNA: 33ng-cm/µl
RNA: 40ng-cm/µl
Absobance measurements are made at A260 and A280. The ratio of absorbance at 260 and 280nm is used to assess the purity of DNA (and RNA). A ratio of 1.8 is accepted as “pure” DNA (2.0 for RNA). Any ratio significantly lower than 1.8 may indicate the presence of protein contaminants. A second ratio of absorbance at 260 and 230nm, with a value between 1.8-2.2 can also indicate good quality DNA.
